What Genetic Makeup Are Most Italians
Abstract
Contempo scientific literature has highlighted the relevance of population genetic studies both for disease clan mapping in admixed populations and for agreement the history of human being migrations. Deeper insight into the history of the Italian population is critical for understanding the peopling of Europe. Because of its crucial position at the centre of the Mediterranean basin, the Italian peninsula has experienced a complex history of colonization and migration whose genetic signatures are still present in contemporary Italians. In this report, we investigated genomic variation in the Italian population using 2.five million unmarried-nucleotide polymorphisms in a sample of more than than 300 unrelated Italian subjects with well-defined geographical origins. We combined several belittling approaches to interpret genome-wide information on 1272 individuals from European, Center Eastern, and North African populations. We detected 3 major bequeathed components contributing dissimilar proportions across the Italian peninsula, and signatures of continuous cistron flow within Italia, which have produced remarkable genetic variability amid contemporary Italians. In addition, we have extracted novel details almost the Italian population'due south beginnings, identifying the genetic signatures of major historical events in Europe and the Mediterranean basin from the Neolithic (e.chiliad., peopling of Sardinia) to recent times (e.1000., 'barbarian invasion' of Northern and Central Italy). These results are valuable for further genetic, epidemiological and forensic studies in Italy and in Europe.
Introduction
The Italian population has a greater degree of internal genomic variability compared with other European countries.one This reflects geographic isolation within Italia, because of its mountainous topography, and to historical events that triggered demographic changes. An example of the latter is the long history of the Roman Empire (27 BCE–476 CE), whose decline was accompanied by a considerable reduction in population size and a series of subsequent migratory waves across Italy. Due to its crucial position at the centre of the Mediterranean basin, the Italian peninsula has experienced a complex history of colonization and migration and the genetic signatures of these human origins are still present in gimmicky Italians.2, 3, 4 Deeper insight into the history of the Italian population is besides critical for understanding the peopling of Europe.
The genetic structure of Italy, whose unity of people and civilisation is quite contempo, was initially analysed using classical genetic markers by Piazza et al. five and Cavalli-Sforza et al. 6 Recently, using genome-wide data, O'Dushlaine et al. 7 defined fine-scale genetic differences among people from different rural villages in Northern and Primal Italy. Geographical patterns of Y chromosomal and mtDNA variety in Italy, mainly determined by the combined action of migrate and founder furnishings, have been described.8, 9 A correlation betwixt genetic and geographic structure in Europe has also been found,10, 11 with a detectable distinction between Southern Italians and other Europeans.
In add-on to evolutionary history, the identification of genetic substructure in manifestly homogeneous populations can better association mapping in admixed populations.12 Moreover, it has recently been shown that variation in susceptibility to certain diseases, and response to drugs or therapies, is related to different proportions of not-European ancestry in admixed populations.thirteen
The aim of this study was to investigate fine-scale Italian population genetic substructure. We used both the large single-nucleotide polymorphism (SNP) data fix, which we collected from a well-characterized Italian sample, and the near recent haplotype-based population genetic algorithms, such as fineSTRUCTURE,xiv which are able to provide finer resolution of the genetic structure of populations, as was shown for the UK population by Leslie et al. 15 Specifically, our aims were to test the feasibility of identifying differences at the microregional level within Italy, to compare and quantify the contributions of populations from Europe and the Mediterranean basin to the genetic composition of the Italians and to explore the historical events that led to the observed high genomic variability within Italia. Several analytical approaches were combined to obtain a consummate portrait of 'the Italian genome', and to exam the robustness of our results across different methodologies. We too traced the major historical events that led to the complex genomic mosaic observed inside Italy, including demographic changes and waves of migration beyond Europe and the Mediterranean basin.
Materials and methods
Subjects
Italian subjects were selected from eleven out of the 20 Italian administrative regions according to a listing of specific region/geographical area/province typical surnames on the basis of previous works by Zei et al. 16, 17 Each subject included in the written report had a well-defined geographical origin: 4 grandparents (and parents) born in the same authoritative region, assessed through interviews at the time of blood collection. The distribution of the study samples across Italia and the sampling provinces is shown in Effigy i and Supplementary Tabular array S1 with sample sizes. Informed consent was provided by all the participants at the time of enrolment. An internal ethical review board at the Homo Genetics Foundation (HuGeF, Comitato Etico HUGEF/15-12-2011) approved this study.
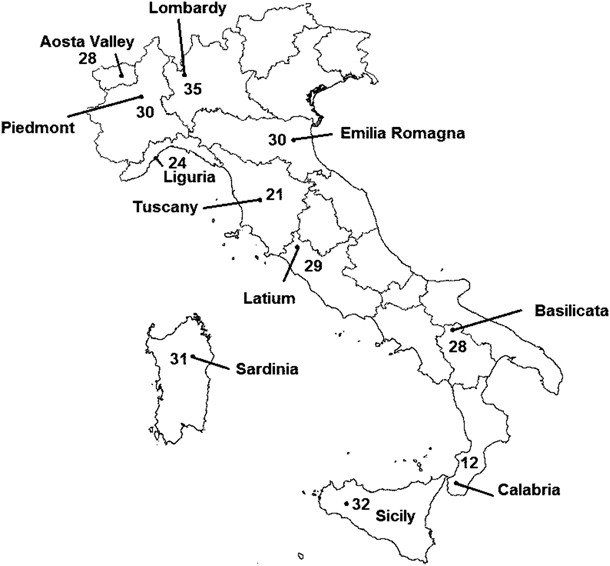
Sampling location barycentre (average sampling place weighted by the number of individuals from each sampling signal) and sample size for each of the 11 Italian regions analysed in this study (Latitude and Longitude position within parentheses): Aosta Valley (45°lxx′N–7°30′E); Basilicata (40°69′N–sixteen°57′E); Calabria (38°10′N–15°70′E); Emilia Romagna (44°80′N–11°60′East); Latium (42°40′N—12°10′E); Liguria (44°30′N–8°l′E; Lombardy (45°39′North–9°85′E); Piedmont (45°19′N–7°90′Due east); Sardinia (40°22′N– 9°xxx′E); Sicily (37°41′N–13°72′East); Tuscany (43°31′N–11°35′East).
Information sets
Two information sets were used to address the different topics of this study.
The first comprised ane 698 926 SNPs investigated in 300 Italian samples, and was used to compare genetic profiles among subjects sampled from the different Italian regions and macroareas. Genome-wide genotype data and the macroarea of origin for each sample volition exist fabricated available via the European Genome-phenome Archive (EGA) repository (https://www.ebi.ac.uk/ega) under the accession number EGAS00001001458, and they are already bachelor upon request from the HuGeF repository (run across Additional information section for the reference spider web link). See Supplementary Methods for details about data drove, Dna extraction, genotyping, quality controls and SNP imputation procedures. The second data gear up comprised 347 131 SNPs assayed on 1272 samples (Tabular array 1), and was used for the comparing between the Italian and non-Italian populations.
Italian population substructure
The Italian population substructure was investigated with the model-based Bayesian cluster algorithm implemented in the combined software ChromoPainter-fineSTRUCTURE,14 using a model that takes into account linkage disequilibrium (LD) between SNPs ('linked' model) with the default parameters. The results were reported as a heatmap (coancestry matrix) of pairwise similarity betwixt subjects, expressed as the number of genomic segments inherited by the same source population. Principal component assay (PCA) was carried out using the same coancestry matrix, and a dendrogram based on hierarchical clustering was constructed. The clustering algorithm uses a Bayesian approach, in which the number of donor populations G and the expected proportion of chunks from each donor population to each individual are inferred by maximum likelihood. Consignment to each cluster is then performed using a Markov Concatenation Monte Carlo method. Run into Lawson et al. 14 for details.
Identical-by-descent calling
Pairwise shared identical-by-descent (IBD) segments were identified by the fastIBD method implemented in BEAGLE v.iii.2eighteen using default parameters. The postprocessing procedure suggested by Ralph and Coop19 was used to minimize the number of imitation-positive calls. Briefly, the IBD call algorithm was run 10 times with 10 unlike random seeds; any segment not overlapping a segment seen in at least ane other run was removed; any 2 segments separated past a gap shorter than at least one of the segments and no more than than v cM long were merged; whatever merged segment that did not contain a subsegment with score below ane × x−9 was removed.
The statistics Due west AB defined by Atzmon et al. 20 and L AB defined past Botigue et al. 21 were computed equally a summary of IBD sharing. The outset is an index of the total length of the shared IBD blocks averaged over the number of possible pairs of individuals (one from population A and the other from population B); the 2d is an alphabetize of the average length of a segment shared IBD between a pair of individuals normalized over the possible number of pairwise comparisons between populations. A cake jackknife procedure was used to compute standard errors and conviction intervals for both estimates.
Isolation-past-distance and genetic boundary tests
The Mantel test22 was applied to the correlation betwixt geographical distance (the shortest distance on a roadmap) expressed in kilometres (km) and genetic similarity measured by the W statistic defined to a higher place. Statistical significance was evaluated by computing an empirical P-value based on 10 000 permutations. To verify the presence of genetic boundaries across the Italian regions, we used Monmonier's algorithm.23 Statistical significance of the genetic boundaries was computed as suggested in Manni et al. 23 Briefly, the test is based on assay of resampled bootstrap matrices: a score is associated with all the dissimilar edges that establish barriers and indicates how many times each edge is included in 1 of the N boundaries computed to determine an empirical P-value.
Effective population size
The effective population size (Northward e) was inferred for each of the 3 Italian macroareas (Northern, Cardinal and Southern Italy) and each of the eleven Italian regions separately, taking advantage of the human relationship between the number and length of shared IBD segments and Due north e as described by Palamara and Pe'er.24 Specifically, nosotros estimated the 'ancestral' N e from the number of short (<2 cM) IBD shared segments but.
Admixture analysis
The unsupervised algorithm implemented in the software ADMIXTURE i.2325 was used to infer ancestry proportions for all the Italian, European, Middle Eastern and North African samples. Nine ancestral cluster arrangements (K=2,…,10) were tested, and the cross- validation fault procedure implemented in the same software was used to define the most realistic contributions of bequeathed populations to the currently observed pattern. The process was repeated 20 times and the results were averaged. To avoid bias due to differences in sample sizes among populations, this assay was performed with a balanced sample size (20 individuals per population). Results were further validated using an independent methodology implemented in the software RFMIX.26 Meet Supplementary Methods for details.
Fst genetic distance
The genetic altitude between all population pairs was assessed by computing the Fst (fixation index) measure, equally implemented in the R packet snpStats.27 Confidence intervals were obtained by means of a block jackknife procedure.
Time since admixture events
To estimate the time of admixture events, we used the extension of the ROLLOFF method implemented in the software ALDER.28 For this assay, the Italian regions were grouped according to the 5 previously identified clusters (Northern, Central, Southern Italy, Aosta Valley and Sardinia), and admixture was tested confronting each of the not-Italian populations included in the written report. See Supplementary Methods for details.
Results
Italian population substructure
The fineSTRUCTURE coancestry matrix is shown in Figure 2a. The pie charts on the bottom panel show the overlap between the observed clusters and the regions of origin. The sensitivity of the cluster algorithm in assigning each sample to the right macroarea was 96.43%, 86.55%, 92.00%, 94.44% and 100% for Aosta Valley, Northern Italia, Central Italia, Southern Italy and Sardinia, respectively, whereas the specificity was 98.16%, 96.68%, 94.eighty%, 99.56% and 100% respectively. In view of the to a higher place results, the Italian regions were divided into v groups for the statistical analyses: Northern, Central, Southern Italy, Aosta Valley and Sardinia.
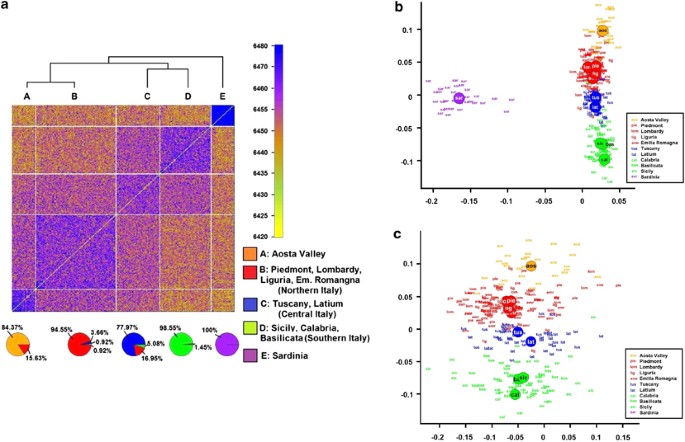
Heatmap representing the coancestry matrix indicating the number of genomic segments inherited from the same ancestral populations for each pair of samples: sendrogram based on hierarchical clustering (at the top), and pie charts representing the overlap between inferred and cocky-reported origin of the Italian individuals (a). Main component analysis based on the aforementioned coancestry matrix including Sardinians (b) and excluding Sardinians (c); 10 and y axes were inverted to emphasize similarity to the geographical map of Italia.
The distribution of the Italian individuals from the commencement 2 eigenvectors of the PCA is shown in Effigy 2, both including and excluding Sardinians (Figures 2b and c). The PCA results provided evidence of large differences between Sardinians and other Italians, and the presence of a genetic gradient across mainland Italia. By performing a genome-broad scan using the start four eigenvectors as contained outcomes regressed against each SNP used every bit a predictor, nosotros establish that the kickoff four PCs strongly correlated with well-known loci under selective pressure, such as the HLA-A complex (hg19 chr6:g.21 266 925_32 628 428),29 and the locus related to the lactase persistence phenotype (hg19 chr2:g.135 907 088_137 013 606);thirty and loci with a low recombination rate such as the hg19 chr8:g.viii 094 406_11 860 625 region harbouring a known polymorphic inversion in Europeans31 (see Manhattan plot – Supplementary Figure S1). In view of the above, nosotros repeated the analysis excluding the in a higher place loci, obtaining the aforementioned pattern of variability across mainland Italy (Supplementary Effigy S2): the correlation between the outset two PCs computed with and without these loci was >0.95. ADMIXTURE assay was also performed on the 300 Italians: meet Supplementary Fabric and Supplementary Effigy S3 for a clarification of the results.
Shared IBD haplotypes across Italia
Nosotros establish greater sharing of IBD segments within regions than between regions from both the Due west and L statistics (Supplementary Tables S2A and S2B), although in some cases the differences were not statistically meaning (data not shown). The Mantel test showed a significant correlation betwixt the geographical distance and the full length of shared IBD segments, both when including Sardinia (R=–0.483, P=0.0039), and when excluding Sardinia from the analysis (R=–0.622, P=0.0027). Based on Monmonier's algorithm, a genetic barrier was identified betwixt Sardinia and mainland Italy (empirical P<0.0001), whereas there was no evidence of any statistically meaning genetic barrier within the peninsula.
A significant s trend of increasing population size North e (Supplementary Effigy S4A) was found (P for trend<0.0001) when the mainland Italians were grouped co-ordinate to the three master macroareas: Northern, Fundamental and Southern. From unmarried region results (Supplementary Figure S4B), the lowest estimated ancestral N e values were for Sardinia and the Aosta Valley (<5000), accompanied by a loftier rate of inbreeding, whereas the effective population sizes were rather homogeneous across the remaining regions.
Comparison with neighbouring populations
We outset used PCA (Supplementary Figure S5) to investigate genetic differences beyond 35 populations from Europe, the Middle East and North Africa. The projection of the offset ii eigenvectors reflects well the geographical origins of the subjects included in the assay (see Supplementary Results for details). We farther investigated population construction using ADMIXTURE. Three major components are noticeable in the Italian population, with different proportions among the major Italian macroareas (see Supplementary Results and Supplementary Effigy S6 for a more detailed description).
The distribution of the pairwise Fst distances between all population pairs is shown in Supplementary Table S3. The genetic altitude between Southern and Northern Italians (Fst=0.0013) is comparable to that between individuals living in different political units (ie, Iberians-Romanians Fst=0.0011; British-French Fst=0.0007), and, interestingly, in >50% of all the possible pairwise comparisons within Europe (Supplementary Figure S7).
Finally, when comparing IBD segment sharing between Italians and the other populations, both with full length West and the average length L, we observed that Southern Italians share more IBD with North African and Middle Eastern populations, whereas Northern Italians share more IBD with Europeans, as is predictable by geography (see Supplementary Results and Supplementary Table S4 for details).
Fourth dimension since admixture events
We estimated time since admixture events from LD disuse equally a function of genetic distance.
Nosotros found evidence of the presence of a mix of Central-Northern European and Center Eastern-Northward African ancestries in the Italian individuals (Supplementary Tabular array S5). The estimated times of admixture ranged between ~2050 and 1300 years ago (y.a.), with an average of most 1650 y.a. – assuming 29 years per generation32– for Northern Italians, and between ~3000 and 1450 y.a. (~2100 y.a. on average) for Fundamental Italians. Finally, for the Southern Italian individuals, admixture between European and Northern African-Middle Eastern ancestry was estimated to have occurred about grand y.a. (run across Supplementary Table S5 and Supplementary Results for a complete written report of significant results).
Word
We evaluated fine-calibration genetic differences within the Italian population using a large prepare of SNPs genotyped in more than 300 Italian individuals and compared them with published genotype data for 1272 European, Middle Eastern and North African individuals. Our report focused on only xi of the 20 Italian regions and, in detail, lacks representation from the Eastern part of Italian republic. On the other hand, some of the strengths of this study are the sample selection criteria based on typical surnames and the identify of nascency of the 4 grandparents, which avoids the inclusion of individuals whose origins are different from their identify of birth. In a previous written report,1 we provided a kickoff overview of the genetic composition of Italians, selecting individuals based on the place of birth simply, but we were not able to discriminate betwixt Northern and Primal Italians. We observed that a proportion of individuals born in Northern Italy amassed with Southern Italians. This was explained by the internal migration that occurred during the terminal ii generations, when people from Southern Italian republic left their place of origin looking for better economic opportunities in the N.
Genetic slope beyond mainland Italian republic
Several of our analyses revealed that the genetic structure of Italians varies to a large extent simply that it can be used to assign each mainland Italian to the right macroarea of origin with very good sensitivity and specificity. The few misclassified individuals could exist because of incorrect self-reported origin of mother or grandparents from the maternal line (sampling according to typical surnames makes the paternal line more likely to be correct), or to different origins of the great-grandparents almost whom we accept no information. The PCA, IBD and ancestry analyses revealed a genetic slope across the peninsula that correlates with geography. As the variety gradient in Italian republic remained later on excluding loci under selective forces or with low recombination rate, we may speculate that historical events have a complementary role in explaining the great genomic variability within the Italian population. Fst genetic distance between Northern and Southern Italians is comparable or even college compared with differences observed amid individuals living in different countries, further confirming the high genomic variability within Italy. We also replicated the previously described slope of SNP allelic frequencies on the hg19 chr2:g.135 907 088_137 013 606 locus,30 related to the lactase persistence phenotype, which strongly correlates with the second PC, and which in plow reflects the geographical location.
Continuous gene menstruum or different ancestral populations
We hypothesize two simple historical scenarios leading to the observed genetic variability across Italy: (a) continuous ancient factor flow amplified by isolation-past-altitude in contempo times; (b) different ancestral origins of the main Italian macroareas whose distinguishability has been attenuated past genetic exchange in recent times.
Monmonier'due south algorithm revealed no prove of the presence of genetic barriers across the peninsula. Instead, results from the Mantel test provide testify of a correlation betwixt genetics and geographical distance. The observed higher average length of the segments with shared IBD inside regions compared with those shared between regions (Supplementary Table S2B) suggests recent isolation-by-distance across the wide range of latitude of the Italian peninsula. Moreover, a North to Southward gradient of increasing ancestral N e was inferred for the 3 main macroareas (Northern, Fundamental and Southern), coinciding with increased heterozygosity in Southern Italy. A similar tendency was previously described for the charge per unit of inbreeding and genome-wide similarity across Central Europe,33 and could be interpreted equally a signature of the 'Out of Africa' migration during Palaeolithic, expansions from refugia after the water ice historic period and of ancient Southward-to-North migratory waves that occurred at the times of European colonization by Neolithic farmers. The beginnings and IBD analyses provided evidence of admixture in Italy with 3 major ancestries detected, most represented in Northern Europeans, Southern Europeans and Center Eastern, respectively (with a pocket-sized percentage of a N African component found in South Italy and Sardinia), with dissimilar prevalence across the peninsula. None of these components is fixed in any population, significant that there is a poor fit with a strict admixture model, as assumed by the algorithm used, and supporting a process of continuous factor flow in multiple directions (migratory waves to and from Italy). Co-ordinate to previous studies on the Y chromosome and mtDNA,34, 35 the Eye Eastern beginnings in Southern Italians well-nigh likely originated at the time of the Greek colonization and, with a smaller percentage, of the subsequent Arabic domination,7 whereas in Central-Northern Italy it is possibly because of the admixture of the ethnic residents with Middle Eastern populations spreading from the Caucasus to Cardinal Europe.19, 21, 28, 36 Our results concur with previously published reports describing a possible maritime route of colonization across Europe, including Italian republic,37 although we cannot exclude the occurrence of more recent demographic events leading to a like scenario. Finally, the homogenous ancestral effective population size across Italian regions could exist interpreted every bit reflecting common genetic origins, taking also into business relationship previous considerations, although the same results might also occur in comparing populations without common origins.
Our study supports the notion that genetic variability across Italy is probable to represent continuous cistron flow leading to differences in the proportion of ancestry from dissimilar sources, along with genetic commutation among neighbouring populations (eg, Northern Italian with European countries, Southern Italian with Middle Eastern and North African ones). Previous studies, analysing uniparental markers, found Y-chromosome genetic aperture beyond Italia. This contrasts with a general lack of construction for mitochondrial DNA,2, 4 and with a higher homogeneity for maternal than paternal genetic contributions, suggesting unlike demographic and historical dynamics for females and males in Italian republic.
Sardinia
Among the 11 Italian regions investigated, Sardinia deserves a separate word. Nosotros replicated previously reported results showing the large genetic differences between Sardinians and mainland Italians;29 the occurrence of a genetic barrier between Sardinia and the residual of Italy; the previously described similarity between the ancestries of the Tyrolean iceman and Sardinians;36 and the very large allelic frequency differences betwixt the islanders and the mainland Italians for the SNPs located on chromosome half dozen, in the HLA-A circuitous locus involved in the immune response,38 which may at least partly explicate the increased prevalence in the island of allowed and autoimmune diseases, such as type ane diabetes and multiple sclerosis.
The Aosta Valley region
The Aosta Valley is a small region at the Due north-Western Italian border with France and Switzerland. Its inhabitants showed interesting genetic characteristics. In the PCA (Figures 2b and c and Supplementary Figure S5), Aostans practise not cluster with subjects from the other Northern Italian regions, non fifty-fifty after the inclusion of non-Italian populations in the assay. Moreover, IBD analysis revealed a high level of inbreeding, comparable to the rate observed in Sardinia. Nevertheless, the estimated proportions of ancestry are comparable to those in Piedmont, Liguria, Lombardy and Emilia Romagna. Hence, our results suggest that the observed differences are non because of the effect of long genetic isolation, equally is the case for Sardinians, but are the results of recent isolation of the Valley. These results are consistent with the low number of dissimilar surnames, mostly of French origin, compared with the number of families, as expected in genetic isolates.39
Time since admixture
The overall procedure to gauge fourth dimension since admixture with the ALDER software is strongly bourgeois and is based on the assumption of the simplified model of admixture we used – that is, a unmarried pulse from detached sources. Our previously discussed results favoured the hypothesis of continuous cistron flow across Italy with admixture events that have probable occurred multiple times, and it should be noted that the method used is designed to emphasize the most contempo admixture event.28 Our estimated admixture dates concur with contempo literature and several historical events. For example, our results support the hypothesis of an admixture event that occurred virtually 3000 y.a. involving populations coming from the Caucasus, the Eye East and populations that lived in Key Italy (Tuscany and Latium), as previously reported from analyses of mitochondrial and Y-chromosome Deoxyribonucleic acid,40 and genome-wide data.41 This was interpreted as possible show of the Heart Eastern (Anatolian) origin of the Etruscans (Herodotus' theory). Admixture events introducing Northern-Central European ancestry into Italy were estimated to have occurred during the and so-called 'Migration Period' later the Roman Empire collapsed (476 CE), with the consequent decline in population. After that the 'Barbarian invasions' took place, with migratory waves from Northern-Fundamental Europe to Northern-Central Italy. It may be speculated that the estimated Northern-Central European ancestry in gimmicky Italians is besides the event of subsequent Italian population growth, as previously reported past studies on mitochondrial and Y-chromosome DNA from a genetic isolate in Northern Italy,42 suggesting that Germanics (Lombards in item) settled in Northern Italia during the 'Migration Period' and may have contributed to the foundation of some communities in Northern Italy. Finally, admixture events involving the Southern Italian population were inferred to have occurred about 1000 y.a., coinciding with The Norman conquest of Southern Italy that spanned virtually of the eleventh and 12th centuries and involved many battles and independent conquerors. A much more detailed assay of geographically well-distributed samples from Southern Italy is required to validate our findings, while a big genetic contribution to the isle of Sicily from Greece has previously been estimated.43
Conclusions and future directions
To our knowledge this is the first report to investigate genetic variability within the Italian population using a very large number of SNPs and subjects with well-defined geographical origin. Nosotros used these data to make inferences on population substructure and admixture events in Italia. To accomplish a more than consummate picture of Italian genetic history and composition, future piece of work should include Italian regions not covered in this report (Eastern regions such as Apulia and Veneto) and should compare Italians with other European populations (Greeks in particular). Our report could be useful for farther genetic, epidemiological and forensic studies in Italia, as it may provide a set of valuable healthy controls for genome-wide association studies, and may be useful for identifying ancestral informative markers. It might also assistance to explain the North-South prevalence slope reported in Italy for several types of tumours.44
Accession codes
Accessions
Gene Expression Passenger vehicle
References
-
Di Gaetano C, Voglino F, Guarrera Southward et al: An overview of the genetic structure within the Italian population from genome-wide data. PLoS One 2012; 7: e43759.
-
Boattini A, Martinez-Cruz B, Sarno Due south et al: Uniparental markers in Italy reveal a sexual activity-biased genetic structure and different historical strata. PLoS I 2013; 8: e65441.
-
Brisighelli F, Alvarez-Iglesias V, Fondevila Yard et al: Uniparental markers of contemporary Italian population reveals details on its pre-Roman heritage. PLoS Ane 2012; vii: e50794.
-
Sarno S, Boattini A, Carta Thousand et al: An aboriginal Mediterranean melting pot: investigating the uniparental genetic construction and population history of sicily and southern Italy. PLoS One 2014; nine: e96074.
-
Piazza A, Cappello N, Olivetti E, Rendine S : A genetic history of Italy. Ann Hum Genet 1988; 52: 203–213.
-
Cavalli-Sforza LL, Menozzi P, Piazza A : The History and Geography of Man Genes. Princeton, NJ, USA: Princeton Universty Press, 1994.
-
O'Dushlaine C, McQuillan R, Weale ME et al: Genes predict village of origin in rural Europe. Eur J Hum Genet 2010; 18: 1269–1270.
-
Di Giacomo F, Luca F, Anagnou N et al: Clinal patterns of human Y chromosomal diversity in continental Italian republic and Greece are dominated by migrate and founder effects. Mol Phylogenet Evol 2003; 28: 387–395.
-
Barbujani Yard, Bertorelle Thou, Capitani Yard, Scozzari R : Geographical structuring in the mtDNA of Italians. Proc Natl Acad Sci USA 1995; 92: 9171–9175.
-
Lao O, Lu TT, Nothnagel M et al: Correlation between genetic and geographic structure in Europe. Curr Biol 2008; 18: 1241–1248.
-
Novembre J, Johnson T, Bryc Thou et al: Genes mirror geography within Europe. Nature 2008; 456: 98–101.
-
Shriner D, Adeyemo A, Ramos East, Chen G, Rotimi CN : Mapping of illness-associated variants in admixed populations. Genome Biol 2011; 12: 223.
-
Seedat YK, Brewster LM : What office does African beginnings play in how hypertensive patients respond to certain antihypertensive drug therapy? Proficient Opin Pharmacother 2014; 15: 159–161.
-
Lawson DJ, Hellenthal M, Myers S, Falush D : Inference of population structure using dense haplotype data. PLoS Genet 2012; 8: e1002453.
-
Leslie S, Winney B, Hellenthal G et al: The fine-scale genetic structure of the British population. Nature 2015; 519: 309–314.
-
Zei G, Barbujani M, Lisa A et al: Barriers to gene flow estimated past surname distribution in Italian republic. Ann Hum Genet 1993; 57: 123–140.
-
Boattini A, Lisa A, Fiorani O, Zei G, Pettener D, Manni F : Full general method to unravel ancient population structures through surnames, concluding validation on Italian information. Hum Biol 2012; 84: 235–270.
-
Browning SR, Browning BL : Rapid and accurate haplotype phasing and missing-information inference for whole-genome association studies by employ of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097.
-
Ralph P, Coop K : The geography of recent genetic ancestry across Europe. PLoS Biol 2013; 11: e1001555.
-
Atzmon G, Hao L, Pe'er I et al: Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet 2010; 86: 850–859.
-
Botigue LR, Henn BM, Gravel Due south et al: Gene flow from N Africa contributes to differential human being genetic diversity in southern Europe. Proc Natl Acad Sci United states of america 2013; 110: 11791–11796.
-
Mantel Northward : The detection of disease clustering and a generalized regression arroyo. Cancer Res 1967; 27: 209–220.
-
Manni F, Guerard E, Heyer E : Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected past using Monmonier'south algorithm. Hum Biol 2004; 76: 173–190.
-
Palamara PF, Pe'er I : Inference of historical migration rates via haplotype sharing. Bioinformatics 2013; 29: i180–i188.
-
Falush D, Stephens Chiliad, Pritchard JK : Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 2007; seven: 574–578.
-
Maples BK, Gravel South, Kenny EE, Bustamante CD : RFMix: a discriminative modeling approach for rapid and robust local-beginnings inference. Am J Hum Genet 2013; 93: 278–288.
-
Clayton D . snpStats: SnpMatrix and XSnpMatrix Classes and Methods. R packet version 1160, 2014.
-
Moorjani P, Patterson N, Hirschhorn JN et al: The history of African cistron flow into Southern Europeans, Levantines, and Jews. PLoS Genet 2011; vii: e1001373.
-
Di Gaetano C, Fiorito Grand, Ortu MF et al: Sardinians genetic background explained past runs of homozygosity and genomic regions nether positive option. PLoS One 2014; 9: e91237.
-
Curry A : Archaeology: the milk revolution. Nature 2013; 500: 20–22.
-
Ma J, Amos CI : Investigation of inversion polymorphisms in the homo genome using primary components analysis. PLoS One 2012; 7: e40224.
-
Fenner JN : Cross-cultural interpretation of the human generation interval for utilize in genetics-based population difference studies. Am J Phys Anthropol 2005; 128: 415–423.
-
Abdellaoui A, Hottenga JJ, de Knijff P et al: Population structure, migration, and diversifying option in the Netherlands. Eur J Hum Genet 2013; 21: 1277–1285.
-
Francalacci P, Morelli L, Underhill PA et al: Peopling of iii Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol 2003; 121: 270–279.
-
Semino O, Magri C, Benuzzi G et al: Origin, improvidence, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
-
Sikora M, Carpenter ML, Moreno-Estrada A et al: Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet 2014; 10: e1004353.
-
Paschou P, Drineas P, Yannaki East et al: Maritime route of colonization of Europe. Proc Natl Acad Sci United states 2014; 111: 9211–9216.
-
Gough SC, Simmonds MJ : The HLA region and autoimmune affliction: associations and mechanisms of action. Curr Genom 2007; 8: 453–465.
-
Berton R : Anthroponomie Valdôtaine. Quart, Aoste, Italia: Musumeci, 1988.
-
Brisighelli F, Capelli C, Alvarez-Iglesias V et al: The Etruscan timeline: a recent Anatolian connection. Eur J Hum Genet 2009; 17: 693–696.
-
Pardo-Seco J, Gomez-Carballa A, Amigo J, Martinon-Torres F, Salas A : A genome-wide study of modernistic-day Tuscans: revisiting Herodotus's theory on the origin of the Etruscans. PLoS One 2014; 9: e105920.
-
Boattini A, Sarno Due south, Pedrini P et al: Traces of medieval migrations in a socially stratified population from Northern Italy. Evidence from uniparental markers and deep-rooted pedigrees. Heredity (Edinb) 2014; 114: 155–162.
-
Di Gaetano C, Cerutti Northward, Crobu F et al: Differential Greek and northern African migrations to Sicily are supported by genetic show from the Y chromosome. Eur J Hum Genet 2009; 17: 91–99.
-
Baili P, De Angelis R, Casella I et al: Italian cancer burden by broad geographical area. Tumori 2007; 93: 398–407.
Acknowledgements
We thank AVIS (Italian Association of Voluntary Claret Donors and specifically Dr Domenico Giupponi), all the volunteers participating in this study and Prof. Luca Cavalli-Sforza for his precious contribution to the study design from the early on stages. C ompagnia di San Paolo, HuGeF and the Stanford Centre for Computational, Evolutionary and Homo Genomics supported this report. Boosted data: The listing of all the genetic and epigenetic information sets are attainable upon request at the HuGeF repository, and data to request data download dominance are bachelor at the following spider web link: http://world wide web.hugef-torino.org/site/alphabetize.php?id=286&t=articolo_secondo_livello&m=extra
Author information
Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of involvement.
Boosted information
Supplementary Data accompanies this paper on European Periodical of Homo Genetics website
Supplementary data
Rights and permissions
Near this commodity
Cite this commodity
Fiorito, Chiliad., Di Gaetano, C., Guarrera, S. et al. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur J Hum Genet 24, 1056–1062 (2016). https://doi.org/10.1038/ejhg.2015.233
-
Received:
-
Revised:
-
Accustomed:
-
Published:
-
Issue Date:
-
DOI : https://doi.org/x.1038/ejhg.2015.233
Further reading
Source: https://www.nature.com/articles/ejhg2015233
Posted by: brennensours1947.blogspot.com

0 Response to "What Genetic Makeup Are Most Italians"
Post a Comment